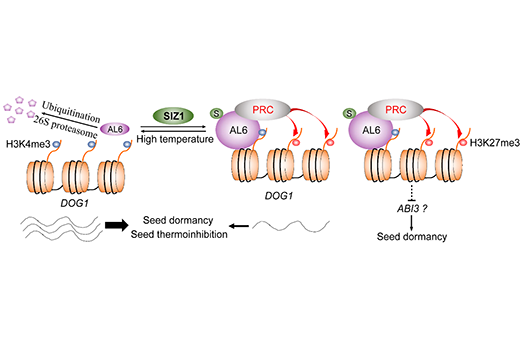
- SUMOylation of AL6 regulates seed dormancy and thermoinhibition in Arabidopsis2024-11-27
- Chromatin Modifier-centered Pathway Points to Higher Crop Yield2024-01-19
- Researchers Discover the Transcriptional Mechanism Underlying Establishment of Callus Pluripotency during in intro Plant Regeneration2023-10-07
- A high-resolution genotype–phenotype map identifies the TaSPL17 controlling grain number and size in wheat2023-08-30
- Scientists Reveal a Molecular Link Between Plant Organ Size Control and Iron Uptake2023-08-14
Publications
more>>
- Bai X, Wu SY, Bai AN, Zhang YM, Zhang Y, Yao XF, Yang T, Chen MM, Liu JL, Li L*, Zhou Y, Liu CM*. 2025. OsSPL9 promotes Cu uptake and translocation in rice grown in high-Fe red soil. New Phytologist, https://doi.org/10.1111/nph.70074 [Detailed content]
- Zhang YY, Zhao HJ, Xiang HX, Zhang JS, Wang L*. 2025. Seasonal and diurnal transcriptome Atlas in natural environment reveals flowering time regulatory network in Alfalfa. Plant Cell & Environment, https://doi.org/10.1111/pce.15466 [Detailed content]
- Deng X#, Pang HY#, Fu Y, Zhang AW, Zhang JY*, Chong K*. 2025. Targeted integrating hyperspectral and metabolomic data with spectral indices and metabolite content models for efficient salt-tolerant phenotype discrimination in Medicago truncatula. Plant Phenomics, https://www.sciencedirect.com/science/article/pii/S2643651525000263 [Detailed content]
- Feng YX, Wang YM, Wang T*, Liu LT*. 2025. NUCLEAR RNA POLYMERASE D1 is essential for tomato embryogenesis and desiccation tolerance in seeds. Cell Reports, https://doi.org/10.1016/j.celrep.2025.115345 [Detailed content]
- He S, Wang XL, Kim S, Kong L, Liu AL, Wang L, Wang Y, Shan LB, He P, Jang J*. 2024. Modulation of stress granule dynamics by phosphorylation and ubiquitination in plants. iSience, 27(11): 111162 [Detailed content]
- He SL#, Li B#, Zahurancik WJ, Arthur HC, Sidharthan V, Gopalan V, Wang L*, Jang JC*. 2024. Overexpression of stress granule protein TZF1 enhances salt stress tolerance by targeting ACA11 mRNA for degradation in Arabidopsis. Frontiers in Plant Science, 15:1375478 [Detailed content]
- Chawla S*, O'Neill J, Knight MI, He YQ, Wang L, Maronde E, Rodríguez SG, van Ooijen G, Garbarino-Pico E, Wolf E, Dkhissi-Benyahya O, Nikhat A, Chakrabarti S, Youngstedt SD, Zi-Ching Mak N, Provencio I, Oster H, Goel N, Caba M, Oosthuizen M, Duffield GE, Chabot C, Davis SJ. 2024. Timely Questions Emerging in Chronobiology: The circadian clock keeps on ticking. Journal of Circadian Rhythms, 22:2. doi: 10.5334/jcr.237 [Detailed content]
- Liu H, Zhao H, Zhang Y, Li X, Zuo Y, Wu Z, Jin K, Xian W, Wang W, Ning W, Liu Z, Zhao X, Wang L, Sage RF, Lu T, Stata M*, Cheng SF*. 2024. The genome of Eleocharis vivipara elucidates the genetics of C3-C4 photosynthetic plasticity and karyotype evolution in the Cyperaceae. Journal of Integrative Plant Biology, 66(11):2505-2527 [Detailed content]
- Guo XY, Luo W, Chong K*. 2024. Exploring abiotic stress resilience module for molecular design in rice. Science Bulletin, doi: https://doi.org/10.1016/j.scib.2024.11.041 [Detailed content]
- Zhao P#, Liu YX#, Deng ZY, Liu LT, Yu TW, Ge GG, Chen BT, Wang T*. 2024. Creating of novel Wx allelic variations signiffcantly altering Wx expression and rice eating and cooking quality. Journal of Plant Physiology, 303: 154384 [Detailed content]