A high-resolution genotype–phenotype map identifies the TaSPL17 controlling grain number and size in wheat
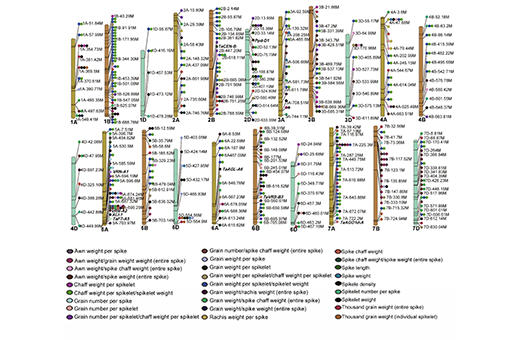
Genotype–phenotype map for the 27 spike morphology traits
Large-scale genotype-phenotype association studies of crop germplasm are important for identifying alleles associated with favorable traits. The limited number of single nucleotide polymorphisms (SNPs) in most wheat genome-wide association studies (GWASs) restricts their power to detect marker-trait associations. Additionally, only a few genes regulating grain number per spikelet have been reported due to sensitivity of this trait to variable environments. We perform a large-scale GWAS using approximately 40 million filtered SNPs for 27 spike morphology traits. We detect 132,086 significant marker-trait associations and the associated SNP markers are located within 590 associated peaks. We detect additional and stronger peaks by dividing spike morphology into sub-traits relative to GWAS results of spike morphology traits. We propose that the genetic dissection of spike morphology is a powerful strategy to detect signals for grain yield traits in wheat. The GWAS results reveal that TaSPL17 positively controls grain size and number by regulating spikelet and floret meristem development, which in turn leads to enhanced grain yield per plant. The haplotypes at TaSPL17 indicate geographical differentiation, domestication effects and breeding selection. Our study provides valuable resources for genetic improvement of spike morphology and a fast-forward genetic solution for candidate gene detection and cloning in wheat.
These results entitled ” A high-resolution genotype–phenotype map identifies the TaSPL17 controlling grain number and size in wheat” has been published in Genome Biology on August 28, 2023. (https://genomebiology.biomedcentral.com/articles/10.1186/s13059-023-03044-2).
Yangyang Liu, PhD student, is one of the first authors and Prof. Zifeng Guo is one of the corresponding authors of this article. This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (Grant No.XDA24010104-2), the Agricultural Science and Technology Innovation Program (CAAS-ZDRW202109) and the National Key Research and Development Program (2020YFE0202300, 2022YFF1002904), the National Natural Science Foundation of China (32225038, 31921005, and 32270269), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24020201), and the Hainan Yazhou Bay Seed Lab (Seed Lab (B21HJ0001 and B21HJ0111).
